Using Names Pipes and Process Substitution in Bioinformatics
It’s hard not to fall in love with Unix as a bioinformatician. In a past post I mentioned how Unix pipes are an extremely elegant way to interface bioinformatics programs (and do inter-process communication in general). In exploring other ways of interfacing programs in Unix, I’ve discovered two great but overlooked ways of interfacing programs: the named pipe and process substitution.
Why We Love Pipes and Unix
A few weeks ago I stumbled across a great talk by Gary Bernhardt entitled The Unix Chainsaw. Bernhardt’s “chainsaw” analogy is great: people sometimes fear doing work in Unix because it’s a powerful tool, and it’s easy to screw up with powerful tools. I think in the process of grokking Unix it’s not uncommon to ask “is this clever and elegant? or completely fucking stupid?”. This is normal, especially if you come from a language like Lisp or Python (or even C really). Unix is a get-shit-done system. I’ve used a chainsaw, and you’re simultaneously amazed at (1) how easily it slices through a tree, and (2) that you’re dumb enough to use this thing three feet away from your vital organs. This is Unix.
Bernhardt also has this great slide, and I’m convinced there’s no better way to describe how most Unix users feel about pipes (especially bioinformaticians):

Pipes are fantastic. Any two (well-written) programs can talk to each other in Unix. All of the nastiness and the difficulty of inter-process communication is solved with one character, |. Thanks Doug McIlroy and others. The stream is usually plaintext, the universal interface, but it doesn’t have to be. With pipes, it doesn’t matter if your pipe is tab delimited marketing data, random email text, or 100 million SNPs. Pipes are a tremendous, beautiful, elegant component of the Unix chainsaw.
But elegance alone won’t earn a software abstraction the hearts of thousands of sysadmins, software engineers, and scientists: pipes are fast. There’s little overheard between pipes, and they are certainly a lot more efficient than writing and reading from the disk. In a past article I included the classic Samtools pipe for SNP calling. It’s no coincidence that other excellent SNP callers like FreeBayes make use of pipes: pipes scale well to moderately large data and they’re just plumbing. Interfacing programs this way allows us to check intermediate output for issues, easily rework entire workflows, and even split off a stream with the aptly named program tee.
Where Pipes Don’t Work
Unix pipes are great, but they don’t work in all situations. The classic problem is in a situation like this:
$ program --in1 in1.txt --in2 in2.txt --out1 out1.txt \
--out2 out2.txt > stats.txt 2> diagnostics.stderrMy past colleagues at the UC Davis Bioinformatics Core and I wrote a set of tools for processing next-generation sequencing data and ran into this situation. In keeping with the Unix traditional, each tool was separate. In practice, this was a crucial design because we saw such differences in data quality due to different sequencing library preparation. Having separate tools working together, in addition to being more Unix-y, lead to more power to spot problems.
However, one step of our workflow has two input files and three output files due to the nature of our data (paired-end sequencing data). Additionally, both in1.txt and in2.txt were the results of another program, and these could be run in parallel (so interleaving the pairs makes this harder to run in parallel). The classic Unix pipe wouldn’t work, as we had more than one input and output into a file: our pipe abstraction breaks down. Hacky solutions like using standard error are way too unpalatable. What to do?
Named Pipes
One solution to this problem is to use named pipes. A named pipe, also known as a FIFO (after First In First Out, a concept in computer science), is a special sort of file we can create with mkfifo:
$ mkfifo fqin
prw-r--r-- 1 vinceb staff 0 Aug 5 22:50 fqinYou’ll notice that this is indeed a special type of file: p for pipe. You interface with these as if they were files (i.e. with Unix redirection, not pipes), but they behave like pipes:
$ echo "hello, named pipes" > fqin &
[1] 16430
$ cat < fqin
[1] + 16430 done echo "hello, named pipes" > fqin
hello, named pipesHopefully you can see the power despite the simple example. Even though the syntax is similar to shell redirection to a file, we’re not actually writing anything to our disk. Note too that the [1] + 16430 done line is printed because we ran the first line as a background process (to free up a prompt). We could also run the same command in a different terminal window. To remove the named pipe, we just use rm.
Creating and using two named pipes would prevent IO bottlenecks and allow us to interface the program creating in1.txt and in2.txt directly with program, but I wanted something cleaner. For quick inter-process communication tasks, I really don’t want to use mkfifo a bunch of times and have to remove each of these named pipes. Luckily Unix offers an even more elegant way: process substitution.
Process Substitution
Process substitution uses the same mechanism as named pipes, but does so without the need to actually create a lasting named pipe through clever shell syntax. These are also appropriately called “anonymous named pipes”. Process substitution is implemented in most modern shells and can be used through the syntax <(command arg1 arg2). The shell runs these commands, and passes their output to a file descriptor, which on Unix systems will be something like /dev/fd/11. This file descriptor will then be substituted by your shell where the call to <() was. Running a command in parenthesis in a shell invokes a seperate subprocess, so multiple uses of <() are run in parallel automatically (scheduling is handled by your OS here, so you may want to use this cautiously on shared systems where more explicity setting the number of processes may be preferable). Additionally, as a subshell, each <() can include its own pipes, so crazy stuff like <(command arg1 | othercommand arg2) is possible, and sometimes wise.
In our simple fake example above, this would look like:
$ program --in1 <(makein raw1.txt) --in2 <(makein raw2.txt) \
--out1 out1.txt --out2 out2.txt > stats.txt 2> diagnostics.stderrwhere makein is some program that creates in1.txt and in2.txt in the original example (from raw1.txt and raw2.txt) and outputs it to standard out. It’s that simple: you’re running a process in a subshell, and its standard out is going to a file descriptor (the /dev/fd/11 or whatever number it is on your system), and program is taking input from that. In fact, if we see this process in htop or with ps, it looks like:
$ ps aux | grep program
vince [...] program --in1 /dev/fd/63 --in2 /dev/fd/62 --out1 out1.txt --out2 out2.txt > stats.txt 2> diagnostics.stderrBut suppose you wanted to pass out1.txt and out2.txt to gzip to compress them? Clearly we don’t want to write them to disk, and then compress them, as this is slow and a waste or system resources. Luckily process substitution works the other way too, through >(). So we could compress in place with:
$ program --in1 <(makein raw1.txt) --in2 <(makein raw2.txt) \
--out1 >(gzip > out.txt.gz) --out2 >(gzip > out2.txt.gz) > stats.txt 2> diagnostics.stderrUnix never ceases to amaze me in its power. The chainsaw is out and you’re cutting through a giant tree. But power comes with a cost here: clarity. Debugging this can be difficult. This level of complexity is like Marmite: I recommend not layering it on too thick at first. You’ll hate it and want to vomit. Admittedly, the nested inter-process communication syntax is neat but awkward — it’s not the simple, clearly understandable | that we’re used to.
Speed
So, is this really faster? Yes, quite. Writing and reading to the disk comes at a big price — see latency numbers every programmer should know. Unfortunately I am too busy to do extensive benchmarks, but I wrote a slightly insane read trimming script that makes use of process substitution. Use at your own risk, but we’re using it over simple Sickle/Scythe/Seqqs combinations. One test uses trim.sh, the other is a simple shell script that just runs Scythe in the background (in parallel, combined with Bash’s wait), writes files to disk, and Sickle processes these. The benchmark is biased against process substitution, because I also compress the files via >(gzip > ) in those tests, but don’t compress the others. Despite my conservative benchmark, the difference is striking:
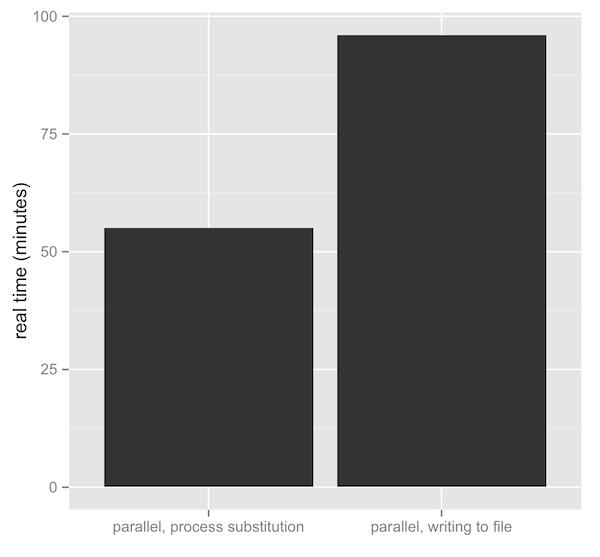
Additionally, with the >(gzip > ) bit, our sequences had a compression ratio of about 3.46% — not bad. With most good tools handling gzip compression natively (that is, without requiring prior decompression), and easy in-place compression via process substitution, there’s really no reason to not keep data large data sets compressed. This is especially the case in bioinformatics where we get decent compression ratios, and our friends less, cat, and grep have their zless, gzcat, and zgrep analogs.
Once again, I’m astonished at the beauty and power of Unix. As far as I know, process substitution is not well know — I asked a few sysadmin friends and they’d seen named pipes but not process substitution. But given Unix’s abstraction of files, it’s no surprise. Actually Brian Kernighan waxed poetically about both pipes and Unix files in this classic AT&T 1980s video on Unix. Hopefully younger generations of programmers will continue to discover the beauty of Unix (and stop re-inventing the wheel, something we’ve all been guilty of). Tools that are designed to work in the Unix environment can leverage Unix’s power end up with emergent powers.
If you want more information on Unix’s named pipes, I suggest:
- Checking out the
teeexample and other examples on Wikipedia’s process substitution page. - This 1997 article from Linux Journal on named pipes.
Share your experiences/wisdom/comments/critiques; I’m on Twitter.